Adapting a classic computer vision algorithm for single cell transcriptomics
This post is about the main idea behind a paper I recently published (and its accompanying software), on a method for aligning scRNA-seq data. Here, aligning means bringing together the representations of two or more datasets so that technical batch effects are reduced. If you were to try analyzing these datasets together without first aligning them, you would probably see them cluster completely apart due to batch effects, despite you expecting them to have a lot of biological similarities.
Imagine you are a robot with a LIDAR sensor. You observe the world around you as “point clouds,” points in a 3D coordinate space representing surfaces. You have two sets of point clouds (“scenes”), each is observing the same environment, but from two different angles.
You could try to operate using only the information from one or the other scene, but then you’d miss out on the additional information about objects that might not be covered by the scene you chose. But if you were to look at them together in the same coordinates, they surfaces wouldn’t line up at all because they have different viewing angles. How would you reconcile the two sets, to unify them into a single, coherent space?
This problem is called “point set registration,” and it is a common problem in computer vision and robotics.
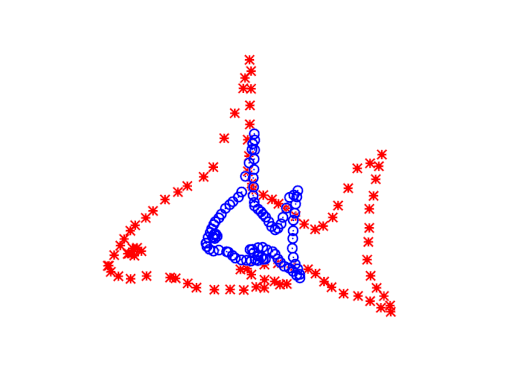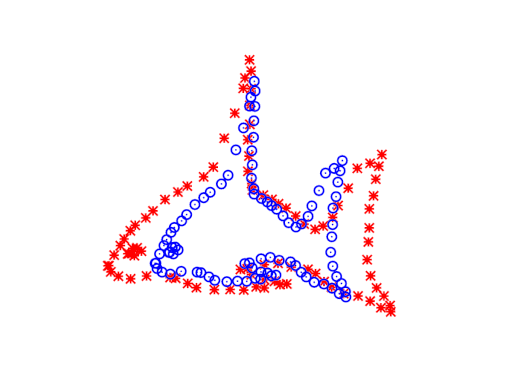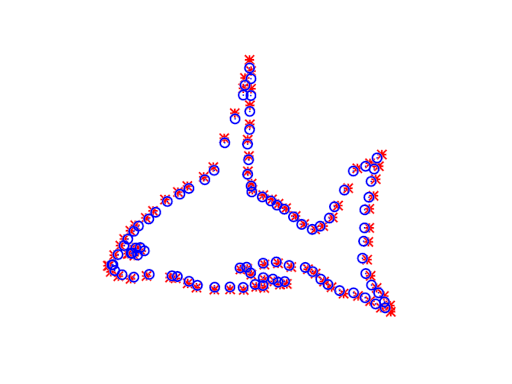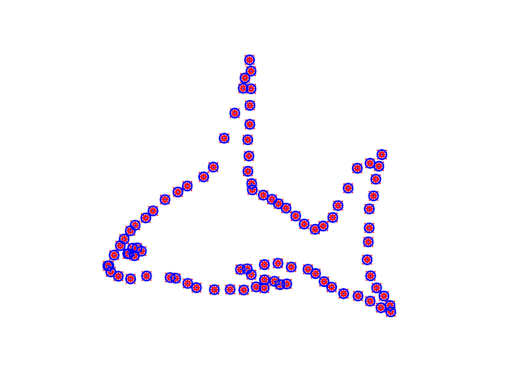
The ICP algorithm
A straightforward approach to aligning these two point clouds is offered by Besl & McKay. Their algorithm, called Iterative Closest Points (ICP), treats one set as a reference or “target” set (let’s call it \(B\)), and holds it fixed while adjusting the other set (\(A\)) to align on top of it. As its name suggests, it’s an iterative algorithm, where each iteration has two steps:
- For each point in \(A\), find the point in \(B\) which is closest to it (euclidean distance)
- Fit a transformation function to apply to all points in \(A\), which reduces the sum of squared distances between the paired-up points from step 1
Then you actually apply the transformation function to the points in \(A\) and repeat the whole process. Various stopping criterion could be used, such as a maximum number of iterations or a threshold of change.
Adapting for scRNA-seq alignment
Now, how can this be useful for scRNA-seq data?
The alignment problem in scRNA-seq has a similar premise: you have two point clouds (data batches), and you believe that they have significant overlap in underlying biological state (for example, they are from the same tissue type, so the cell types should overlap a lot).
However, the key differences here are:
- scRNA-seq point clouds are much much more high-dimensional than LIDAR data (there are often on the order of 20,000 dimensions, one for each gene)
- The assumption in ICP that a rigid transformation is enough to align the point clouds (because they are of rigid objects) is not appropriate for scRNA-seq, as the relationship between genes can be complicated and non-linear, and is often mixed with noise too.
So to adapt ICP to work with scRNA-seq data, we first select only the highly variable genes to look at, reducing the dimensions as much as we can. Then we also change the class of the transformation function, instead using the more flexible family of affine functions, not the overly simple class of rigid transformations.
We call the algorithm Single Cell Iterative Point set Registration (SCIPR). In our paper, we experimented with different methods for pairing up the cells between the sets, as well as different classes of transformation functions. Surprisingly, we found that this fairly simple algorithm, with just affine transformations, was able to align several datasets, with results that were competitive with the state-of-the-art scRNA-seq alignment methods.
A major goal of mine, however, was to release an extensible piece of software around this algorithm, that would serve as a jumping-off point for others to experiment with this iterative procedure. I created a python package, called scipr, with a homepage here. At it’s core, the framework is implemented with the Strategy design pattern from the Gang of Four book, allowing anyone to mix and match different pairing algorithms and transformation functions. We include a slew of our own, but you can easily write your own. Check out the documentation for further explanation.

Comments